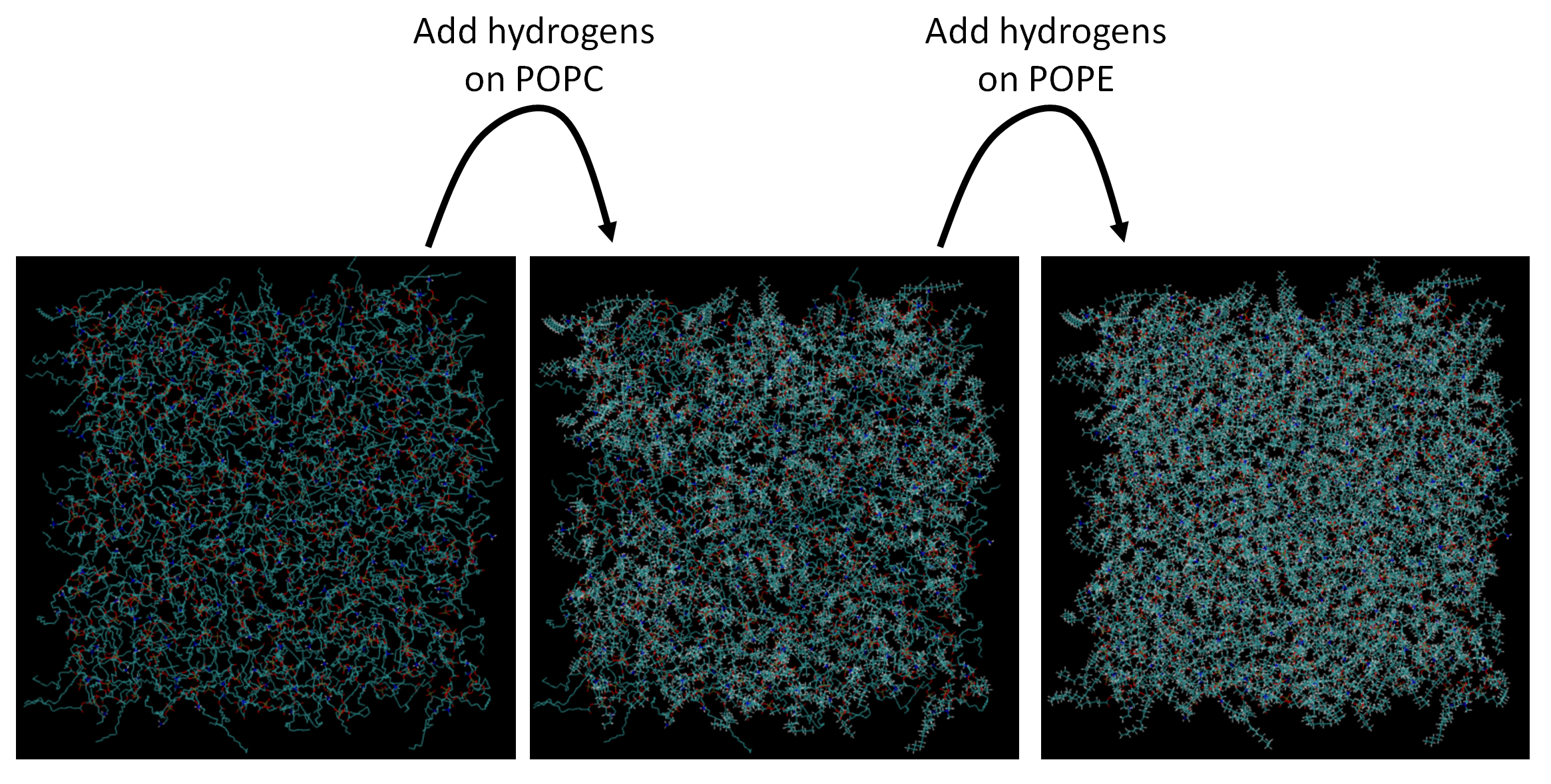
Notebook03: POPC/POPE mixture
Contents
Notebook03: POPC/POPE mixture#
In this notebook, we will show you how to reconstruct hydrogens, calculate the order parameters and produce output trajectories on a POPC/POPE (50:50) mixture. Again, this example is based on the Berger united-atom force field.
Before going on, we advise you to get started with Notebook01 if you are not familiar with basic features of buildH and Jupyter notebooks (e.g. do you know what ! means in a Jupyter cell?).
Checking buildH activation#
As explained in Notebook01, you should have activated buildH before launching this notebook, thus you should obtain the following when invoking buildH in a Unix terminal:
[1]:
!buildH
usage: buildH [-h] [-v] -c COORD [-t TRAJ] -l LIPID
[-lt LIPID_TOPOLOGY [LIPID_TOPOLOGY ...]] -d DEFOP
[-opx OPDBXTC] [-o OUT] [-b BEGIN] [-e END] [-igch3]
buildH: error: the following arguments are required: -c/--coord, -l/--lipid, -d/--defop
If you see this, you can go on. If not, please go to Notebook01 and see the explanations there on how to activate buildH.
Downloading the example files#
The example shown in this Notebook are taken from a POPC/POPE (50:50) trajectory of 256 lipids taken from the NMRlipidsIVb project. The initial source files can be found on Zenodo. For the purpose of learning, we simplified the trajectory and extracted only 25 frames (0-25 ns, one frame / ns). The starting gro file and the trajectory of 25 frames can be directly downloaded
from the buildH repo in the directory https://github.com/patrickfuchs/buildH/tree/master/docs/Berger_POPCPOPE_test_case. Here we’ll download them with wget (recall to download all files in raw format):
[2]:
!wget https://raw.githubusercontent.com/patrickfuchs/buildH/master/docs/Berger_POPCPOPE_test_case/start.gro
--2021-07-07 15:12:13-- https://raw.githubusercontent.com/patrickfuchs/buildH/master/docs/Berger_POPCPOPE_test_case/start.gro
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.110.133, 185.199.111.133, 185.199.108.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.110.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 2097099 (2.0M) [text/plain]
Saving to: ‘start.gro’
start.gro 100%[===================>] 2.00M --.-KB/s in 0.06s
2021-07-07 15:12:13 (32.2 MB/s) - ‘start.gro’ saved [2097099/2097099]
[3]:
!wget https://github.com/patrickfuchs/buildH/raw/master/docs/Berger_POPCPOPE_test_case/md_0_25ns_dt1000.xtc
--2021-07-07 15:12:28-- https://github.com/patrickfuchs/buildH/raw/master/docs/Berger_POPCPOPE_test_case/md_0_25ns_dt1000.xtc
Resolving github.com (github.com)... 140.82.121.4
Connecting to github.com (github.com)|140.82.121.4|:443... connected.
HTTP request sent, awaiting response... 302 Found
Location: https://raw.githubusercontent.com/patrickfuchs/buildH/master/docs/Berger_POPCPOPE_test_case/md_0_25ns_dt1000.xtc [following]
--2021-07-07 15:12:29-- https://raw.githubusercontent.com/patrickfuchs/buildH/master/docs/Berger_POPCPOPE_test_case/md_0_25ns_dt1000.xtc
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.109.133, 185.199.110.133, 185.199.111.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.109.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 4458540 (4.3M) [application/octet-stream]
Saving to: ‘md_0_25ns_dt1000.xtc’
md_0_25ns_dt1000.xt 100%[===================>] 4.25M --.-KB/s in 0.08s
2021-07-07 15:12:29 (56.1 MB/s) - ‘md_0_25ns_dt1000.xtc’ saved [4458540/4458540]
[4]:
!ls
data
img
md_0_25ns_dt1000.xtc
Notebook_01_buildH_calc_OP.ipynb
Notebook_02_buildH_calc_OP_outputwH.ipynb
Notebook_03_mixture_POPC_POPE.ipynb
Notebook_04_library.ipynb
Notebook_05_mixture_POPC_cholesterol.ipynb
start.gro
We also need the two def files corresponding to POPC and POPE. They can be downloaded from the buildH repo in the directory https://github.com/patrickfuchs/buildH/tree/master/def_files (again remember to use raw files):
[5]:
!wget https://raw.githubusercontent.com/patrickfuchs/buildH/master/def_files/Berger_POPC.def
--2021-07-07 15:12:52-- https://raw.githubusercontent.com/patrickfuchs/buildH/master/def_files/Berger_POPC.def
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.111.133, 185.199.108.133, 185.199.109.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.111.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 2135 (2.1K) [text/plain]
Saving to: ‘Berger_POPC.def’
Berger_POPC.def 100%[===================>] 2.08K --.-KB/s in 0.001s
2021-07-07 15:12:53 (2.00 MB/s) - ‘Berger_POPC.def’ saved [2135/2135]
[6]:
!wget https://raw.githubusercontent.com/patrickfuchs/buildH/master/def_files/Berger_POPE.def
--2021-07-07 15:12:56-- https://raw.githubusercontent.com/patrickfuchs/buildH/master/def_files/Berger_POPE.def
Resolving raw.githubusercontent.com (raw.githubusercontent.com)... 185.199.110.133, 185.199.111.133, 185.199.108.133, ...
Connecting to raw.githubusercontent.com (raw.githubusercontent.com)|185.199.110.133|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 1937 (1.9K) [text/plain]
Saving to: ‘Berger_POPE.def’
Berger_POPE.def 100%[===================>] 1.89K --.-KB/s in 0.001s
2021-07-07 15:12:56 (1.98 MB/s) - ‘Berger_POPE.def’ saved [1937/1937]
[7]:
!ls
Berger_POPC.def Notebook_02_buildH_calc_OP_outputwH.ipynb
Berger_POPE.def Notebook_03_mixture_POPC_POPE.ipynb
data Notebook_04_library.ipynb
img Notebook_05_mixture_POPC_cholesterol.ipynb
md_0_25ns_dt1000.xtc start.gro
Notebook_01_buildH_calc_OP.ipynb
Recall, the def files present on the buildH def_files directory contain all the possible (apolar) H that can be reconstructed on each lipids. The polar hydrogens are already present (e.g. the different N-H of the ethanolamine in POPE), thus they do not need to be reconstructed. buildH will let them as is, like all other non apolar H atoms.
Dealing with POPC#
When we have a mixture of lipids, buildH cannot reconstruct hydrogens on all lipids at the same time. So we start with H reconstruction and OP calculation on POPC (we’ll focus on POPE later). Since Berger POPC is a supported lipid, we can launch easily buildH with the classical flags: -c for reading the initial pdb or gro file, -t for reading the trajectory, -l for specifying the type of lipid, -d for specifying the def file, -o for the output file with order
parameters, and last -opx for the output trajectory with hydrogens (recall not to put any extension, they’ll be added automatically by buildH). Importantly, when we want to output a trajectory with newly reconstructed hydrogens (option -opx), all possible H to reconstruct need to be present in the def file. This is fine since this is the case for all lipids in the def file directory of the buildH repository.
[8]:
!buildH -c start.gro -l Berger_POPC -t md_0_25ns_dt1000.xtc -d Berger_POPC.def -o OP_POPC.dat -opx traj_POPC_wH
Constructing the system...
System has 46600 atoms
Writing new pdb with hydrogens.
Writing trajectory with hydrogens in xtc file.
Dealing with frame 0 at 0.0 ps.
Dealing with frame 1 at 1000.0 ps.
Dealing with frame 2 at 2000.0 ps.
Dealing with frame 3 at 3000.0 ps.
Dealing with frame 4 at 4000.0 ps.
Dealing with frame 5 at 5000.0 ps.
Dealing with frame 6 at 6000.0 ps.
Dealing with frame 7 at 7000.0 ps.
Dealing with frame 8 at 8000.0 ps.
Dealing with frame 9 at 9000.0 ps.
Dealing with frame 10 at 10000.0 ps.
Dealing with frame 11 at 11000.0 ps.
Dealing with frame 12 at 12000.0 ps.
Dealing with frame 13 at 13000.0 ps.
Dealing with frame 14 at 14000.0 ps.
Dealing with frame 15 at 15000.0 ps.
Dealing with frame 16 at 16000.0 ps.
Dealing with frame 17 at 17000.0 ps.
Dealing with frame 18 at 18000.0 ps.
Dealing with frame 19 at 19000.0 ps.
Dealing with frame 20 at 20000.0 ps.
Dealing with frame 21 at 21000.0 ps.
Dealing with frame 22 at 22000.0 ps.
Dealing with frame 23 at 23000.0 ps.
Dealing with frame 24 at 24000.0 ps.
Dealing with frame 25 at 25000.0 ps.
Results written to OP_POPC.dat
[9]:
!ls
Berger_POPC.def
Berger_POPE.def
data
img
md_0_25ns_dt1000.xtc
Notebook_01_buildH_calc_OP.ipynb
Notebook_02_buildH_calc_OP_outputwH.ipynb
Notebook_03_mixture_POPC_POPE.ipynb
Notebook_04_library.ipynb
Notebook_05_mixture_POPC_cholesterol.ipynb
OP_POPC.dat
start.gro
traj_POPC_wH.pdb
traj_POPC_wH.xtc
The order parameters of POPC lipids have been written to the file OP_POPC.dat
[10]:
!head OP_POPC.dat
# OP_name resname atom1 atom2 OP_mean OP_stddev OP_stem
#--------------------------------------------------------------------
gamma1_1 POPC C1 H11 -0.00343 0.12015 0.01062
gamma1_2 POPC C1 H12 0.00752 0.09808 0.00867
gamma1_3 POPC C1 H13 -0.00614 0.08702 0.00769
gamma2_1 POPC C2 H21 -0.00397 0.11983 0.01059
gamma2_2 POPC C2 H22 -0.00703 0.09226 0.00816
gamma2_3 POPC C2 H23 -0.00952 0.10068 0.00890
gamma3_1 POPC C3 H31 0.00060 0.12174 0.01076
gamma3_2 POPC C3 H32 -0.00474 0.09119 0.00806
A pdb traj_POPC_wH.pdb and an xtc traj_POPC_wH.xtc file have been produced with hydrogens on POPC only. So let’s check whether the POPC have hydrogens and not POPE:
[11]:
!grep POPC traj_POPC_wH.pdb | head -20
ATOM 1 C1 POPC 1 89.160 7.570 22.770 1.00 0.00 C
ATOM 2 H11 POPC 1 88.713 8.530 23.029 1.00 0.00 H
ATOM 3 H12 POPC 1 88.372 6.843 22.577 1.00 0.00 H
ATOM 4 H13 POPC 1 89.775 7.684 21.878 1.00 0.00 H
ATOM 5 C2 POPC 1 89.200 7.090 25.120 1.00 0.00 C
ATOM 6 H21 POPC 1 88.733 8.065 25.257 1.00 0.00 H
ATOM 7 H22 POPC 1 89.845 6.873 25.971 1.00 0.00 H
ATOM 8 H23 POPC 1 88.428 6.324 25.045 1.00 0.00 H
ATOM 9 C3 POPC 1 91.030 8.130 24.080 1.00 0.00 C
ATOM 10 H31 POPC 1 90.552 9.093 24.258 1.00 0.00 H
ATOM 11 H32 POPC 1 91.651 8.192 23.186 1.00 0.00 H
ATOM 12 H33 POPC 1 91.651 7.869 24.937 1.00 0.00 H
ATOM 13 N4 POPC 1 90.000 7.100 23.890 1.00 0.00 N
ATOM 14 C5 POPC 1 90.660 5.800 23.650 1.00 0.00 C
ATOM 15 H51 POPC 1 91.220 5.539 24.548 1.00 0.00 H
ATOM 16 H52 POPC 1 91.350 5.924 22.815 1.00 0.00 H
ATOM 17 C6 POPC 1 89.730 4.630 23.320 1.00 0.00 C
ATOM 18 H61 POPC 1 89.359 4.775 22.305 1.00 0.00 H
ATOM 19 H62 POPC 1 88.895 4.660 24.020 1.00 0.00 H
ATOM 20 O7 POPC 1 90.310 3.330 23.390 1.00 0.00 O
grep: write error: Broken pipe
[12]:
!grep POPE traj_POPC_wH.pdb | head -20
ATOM 17153 H1 POPE 129 82.600 -2.110 24.640 1.00 0.00 H
ATOM 17154 H2 POPE 129 82.290 -1.570 23.100 1.00 0.00 H
ATOM 17155 H3 POPE 129 83.140 -0.610 24.130 1.00 0.00 H
ATOM 17156 N4 POPE 129 82.400 -1.300 24.070 1.00 0.00 N
ATOM 17157 C5 POPE 129 81.140 -0.680 24.510 1.00 0.00 C
ATOM 17158 C6 POPE 129 80.830 0.480 23.550 1.00 0.00 C
ATOM 17159 O7 POPE 129 81.880 1.450 23.530 1.00 0.00 O
ATOM 17160 P8 POPE 129 81.420 2.670 22.580 1.00 0.00 P
ATOM 17161 O9 POPE 129 82.620 3.440 22.200 1.00 0.00 O
ATOM 17162 O10 POPE 129 80.420 2.190 21.610 1.00 0.00 O
ATOM 17163 O11 POPE 129 80.600 3.490 23.700 1.00 0.00 O
ATOM 17164 C12 POPE 129 81.550 4.230 24.480 1.00 0.00 C
ATOM 17165 C13 POPE 129 80.830 5.150 25.470 1.00 0.00 C
ATOM 17166 O14 POPE 129 81.780 5.780 26.340 1.00 0.00 O
ATOM 17167 C15 POPE 129 82.000 7.120 26.230 1.00 0.00 C
ATOM 17168 O16 POPE 129 81.490 7.750 25.300 1.00 0.00 O
ATOM 17169 C17 POPE 129 83.030 7.710 27.190 1.00 0.00 C
ATOM 17170 C18 POPE 129 82.420 8.040 28.550 1.00 0.00 C
ATOM 17171 C19 POPE 129 81.550 9.300 28.470 1.00 0.00 C
ATOM 17172 C20 POPE 129 80.810 9.530 29.790 1.00 0.00 C
grep: write error: Broken pipe
Yeah that is fine! POPC have all hydrogens reconstructed, while POPE do not (we can only see the polar hydrogens which are natively present in a united-atom structure).
Dealing with POPE#
Now that POPC have all hydrogens, we can restart from that pdb file to reconstruct the hydrogens of POPE. Thus we can use the flags -c with the pdb file we just generated traj_POPC_wH.pdb (Hs on POPC but not POPE). The -t will be used with the same trajectory with H on POPC (but not POPE!) traj_POPC_wH.xtc. We output order parameters for POPE with -o. Last, -opx will produce a file with hydrogens on POPE.
[13]:
!buildH -c traj_POPC_wH.pdb -l Berger_POPE -t traj_POPC_wH.xtc -d Berger_POPE.def -o OP_POPE.dat -opx traj_POPCPOPE_wH
Constructing the system...
System has 57096 atoms
Writing new pdb with hydrogens.
Writing trajectory with hydrogens in xtc file.
Dealing with frame 0 at 0.0 ps.
Dealing with frame 1 at 0.0 ps.
Dealing with frame 2 at 0.0 ps.
Dealing with frame 3 at 0.0 ps.
Dealing with frame 4 at 0.0 ps.
Dealing with frame 5 at 0.0 ps.
Dealing with frame 6 at 0.0 ps.
Dealing with frame 7 at 0.0 ps.
Dealing with frame 8 at 0.0 ps.
Dealing with frame 9 at 0.0 ps.
Dealing with frame 10 at 0.0 ps.
Dealing with frame 11 at 0.0 ps.
Dealing with frame 12 at 0.0 ps.
Dealing with frame 13 at 0.0 ps.
Dealing with frame 14 at 0.0 ps.
Dealing with frame 15 at 0.0 ps.
Dealing with frame 16 at 0.0 ps.
Dealing with frame 17 at 0.0 ps.
Dealing with frame 18 at 0.0 ps.
Dealing with frame 19 at 0.0 ps.
Dealing with frame 20 at 0.0 ps.
Dealing with frame 21 at 0.0 ps.
Dealing with frame 22 at 0.0 ps.
Dealing with frame 23 at 0.0 ps.
Dealing with frame 24 at 0.0 ps.
Dealing with frame 25 at 0.0 ps.
Results written to OP_POPE.dat
[14]:
!ls
Berger_POPC.def
Berger_POPE.def
data
img
md_0_25ns_dt1000.xtc
Notebook_01_buildH_calc_OP.ipynb
Notebook_02_buildH_calc_OP_outputwH.ipynb
Notebook_03_mixture_POPC_POPE.ipynb
Notebook_04_library.ipynb
Notebook_05_mixture_POPC_cholesterol.ipynb
OP_POPC.dat
OP_POPE.dat
start.gro
traj_POPCPOPE_wH.pdb
traj_POPCPOPE_wH.xtc
traj_POPC_wH.pdb
traj_POPC_wH.xtc
Great, we can now look at the order parameters of POPE:
[15]:
!head OP_POPE.dat
# OP_name resname atom1 atom2 OP_mean OP_stddev OP_stem
#--------------------------------------------------------------------
beta1 POPE C5 H51 0.18091 0.44608 0.03943
beta2 POPE C5 H52 -0.04410 0.44630 0.03945
alpha1 POPE C6 H61 0.08958 0.45722 0.04041
alpha2 POPE C6 H62 0.19382 0.46467 0.04107
g3_1 POPE C12 H121 -0.27276 0.26619 0.02353
g3_2 POPE C12 H122 -0.18926 0.29892 0.02642
g2_1 POPE C13 H131 -0.15531 0.32935 0.02911
g1_1 POPE C32 H321 0.12967 0.45277 0.04002
And check whether POPC and POPE have all hydrogens reconstructed:
[16]:
!grep POPE traj_POPCPOPE_wH.pdb | head -20
ATOM 17153 H1 POPE 129 82.600 -2.110 24.640 1.00 0.00 H
ATOM 17154 H2 POPE 129 82.290 -1.570 23.100 1.00 0.00 H
ATOM 17155 H3 POPE 129 83.140 -0.610 24.130 1.00 0.00 H
ATOM 17156 N4 POPE 129 82.400 -1.300 24.070 1.00 0.00 N
ATOM 17157 C5 POPE 129 81.140 -0.680 24.510 1.00 0.00 C
ATOM 17158 H51 POPE 129 81.247 -0.302 25.527 1.00 0.00 H
ATOM 17159 H52 POPE 129 80.334 -1.414 24.478 1.00 0.00 H
ATOM 17160 C6 POPE 129 80.830 0.480 23.550 1.00 0.00 C
ATOM 17161 H61 POPE 129 79.908 0.965 23.870 1.00 0.00 H
ATOM 17162 H62 POPE 129 80.700 0.080 22.544 1.00 0.00 H
ATOM 17163 O7 POPE 129 81.880 1.450 23.530 1.00 0.00 O
ATOM 17164 P8 POPE 129 81.420 2.670 22.580 1.00 0.00 P
ATOM 17165 O9 POPE 129 82.620 3.440 22.200 1.00 0.00 O
ATOM 17166 O10 POPE 129 80.420 2.190 21.610 1.00 0.00 O
ATOM 17167 O11 POPE 129 80.600 3.490 23.700 1.00 0.00 O
ATOM 17168 C12 POPE 129 81.550 4.230 24.480 1.00 0.00 C
ATOM 17169 H121 POPE 129 82.169 4.833 23.815 1.00 0.00 H
ATOM 17170 H122 POPE 129 82.181 3.534 25.032 1.00 0.00 H
ATOM 17171 C13 POPE 129 80.830 5.150 25.470 1.00 0.00 C
ATOM 17172 H131 POPE 129 80.311 5.977 24.987 1.00 0.00 H
grep: write error: Broken pipe
[17]:
!grep POPC traj_POPCPOPE_wH.pdb | head -20
ATOM 1 C1 POPC 1 89.160 7.570 22.770 1.00 0.00 C
ATOM 2 H11 POPC 1 88.710 8.530 23.030 1.00 0.00 H
ATOM 3 H12 POPC 1 88.370 6.840 22.580 1.00 0.00 H
ATOM 4 H13 POPC 1 89.780 7.680 21.880 1.00 0.00 H
ATOM 5 C2 POPC 1 89.200 7.090 25.120 1.00 0.00 C
ATOM 6 H21 POPC 1 88.730 8.060 25.260 1.00 0.00 H
ATOM 7 H22 POPC 1 89.850 6.870 25.970 1.00 0.00 H
ATOM 8 H23 POPC 1 88.430 6.320 25.050 1.00 0.00 H
ATOM 9 C3 POPC 1 91.030 8.130 24.080 1.00 0.00 C
ATOM 10 H31 POPC 1 90.550 9.090 24.260 1.00 0.00 H
ATOM 11 H32 POPC 1 91.650 8.190 23.190 1.00 0.00 H
ATOM 12 H33 POPC 1 91.650 7.870 24.940 1.00 0.00 H
ATOM 13 N4 POPC 1 90.000 7.100 23.890 1.00 0.00 N
ATOM 14 C5 POPC 1 90.660 5.800 23.650 1.00 0.00 C
ATOM 15 H51 POPC 1 91.220 5.540 24.550 1.00 0.00 H
ATOM 16 H52 POPC 1 91.350 5.920 22.820 1.00 0.00 H
ATOM 17 C6 POPC 1 89.730 4.630 23.320 1.00 0.00 C
ATOM 18 H61 POPC 1 89.360 4.780 22.310 1.00 0.00 H
ATOM 19 H62 POPC 1 88.900 4.660 24.020 1.00 0.00 H
ATOM 20 O7 POPC 1 90.310 3.330 23.390 1.00 0.00 O
grep: write error: Broken pipe
Conclusion#
In this notebook, we showed you how to use buildH on a mixture of POPC/POPE lipids. buildH has to be launched for each lipid separately. If a trajectory with all hydrogens on all lipids is wanted, one has to produce intermediate trajectories at each step. In the figure below is shown the two step process (image rendered with VMD).